News release
From:
Biology: Creating atlases of all human cells (N&V) *PRESS BRIEFING* *IMAGES, VIDEO & GIF*
Early draft maps of cells in the human body are described in a collection of papers from the Human Cell Atlas (HCA) initiative, published across the Nature Portfolio journals. The research leverages new data and analytical tools, some based on artificial intelligence and machine learning, to aid our understanding of human health and disease at a cellular level.
The human body comprises an estimated 37.2 trillion cells, and each cell type has a unique function. Understanding the complexity of the human body at the cellular level has been challenging, but is crucial for the advancement of medical science. The HCA consortium was founded in 2016 with the aim of building a biological atlas of every cell type in the human body; the consortium consists of more than 3,600 members in 102 countries, who contribute data related to 18 biological networks.
The latest collection highlights recent findings in three key areas from the consortium. First, new data have been generated from human developmental tissues; for example, Sarah Teichmann, Ken To and colleagues provide new data for skull bones and hip, knee and shoulder joints. Second, the consortium has developed analytical tools, including a machine learning-based method that searches for similar cells on the basis of their expression profiles. Third, the collection features integrated analyses of data that are available for specific organs or biological systems.
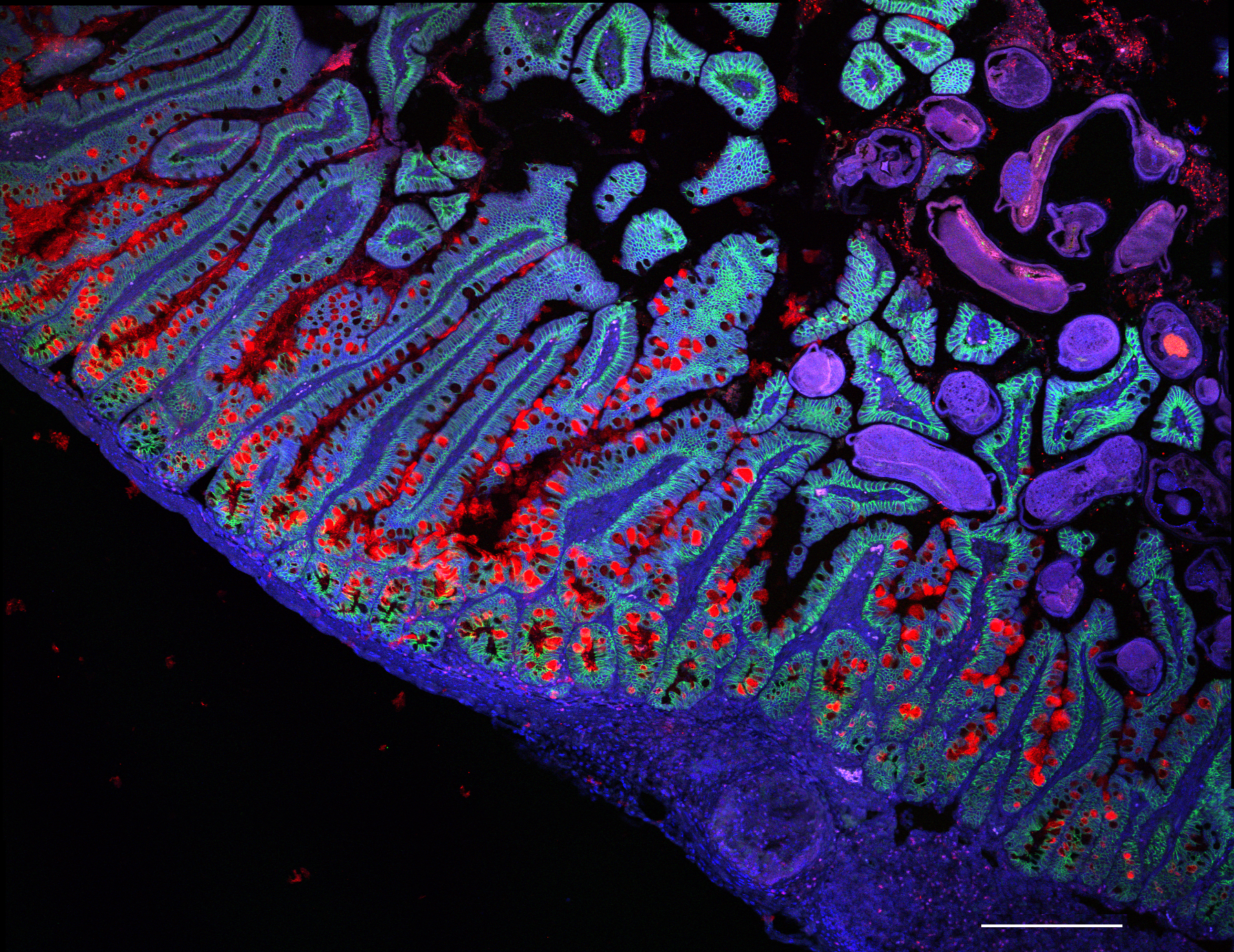
For example, Amanda Oliver and colleagues present the gastrointestinal tract atlas, which spans from tissues of the mouth through to the oesophagus, stomach, intestines and colon, and also includes data from individuals with inflammatory diseases such as Crohn’s disease. Additionally, an integrated brain organoid cell atlas, developed by Barbara Treutlein and colleagues, provides insights into how well organoids capture aspects of the developing brain.
Together, these findings represent important advances in the construction of the first HCA cell draft map, which will have many potential future implications. For example, it will improve our understanding of how cellular diversity influences individual responses to medical treatments and help with investigating the genetic underpinnings of diseases at a cellular level. The insights gained from the maps are discussed in an accompanying Perspective by Aviv Regev and colleagues.
Although challenges remain in fully capturing the dynamic nature of cells and extending these insights across diverse populations, the ongoing collaboration among scientists worldwide will help to make personalized medicine more attainable and enhance our ability to treat diseases.
Multimedia




 Australia; International; NSW
Australia; International; NSW