News release
From:
Australian researchers have developed a powerful new way to target deadly, drug-resistant bacteria by designing antibodies that recognise a sugar found only on bacterial cells – an advance that could underpin a new generation of immunotherapies for multidrug resistant hospital-acquired infections.
Published in Nature Chemical Biology, the research shows that a laboratory-made antibody can clear an otherwise lethal bacterial infection in mice by homing in on a distinctive bacterial sugar and flagging the pathogen for destruction by the immune system.
The work was co-led by Professor Richard Payne from the University of Sydney, in collaboration with Professor Ethan Goddard-Borger at WEHI and Associate Professor Nichollas Scott at the University of Melbourne and the Peter Doherty Institute for Infection and Immunity.
Professor Payne will also lead the recently announced Australian Research Council Centre of Excellence for Advanced Peptide and Protein Engineering, which will build on discoveries like this to accelerate translation into applications in biotechnology, agriculture and conservation.
“This study shows what’s possible when we combine chemical synthesis with biochemistry, immunology, microbiology and infection biology,” Professor Payne said. “By precisely building these bacterial sugars in the lab with synthetic chemistry, we were able to understand their shape at the molecular level and develop antibodies that bind them with high specificity. That opens the door to new ways of treating some devastating drug-resistant bacterial infections.”
The target of the new antibody is a sugar molecule known as pseudaminic acid. While it resembles other sugars found on human cells, it is produced exclusively by bacteria and is used by many dangerous pathogens as essential components of their outer coats and evade immune responses.
Because humans do not make this sugar, it represents a highly differentiated target for immunotherapy development.
To exploit this vulnerability, the team first chemically synthesised the bacterial sugar and sugar-decorated peptides from scratch. This allowed them to determine the exact three-dimensional arrangement of the molecule and how it is presented on bacterial surfaces. Using these insights and molecules, they developed a “pan-specific” antibody capable of recognising the sugar across a wide range of bacterial species and strains.
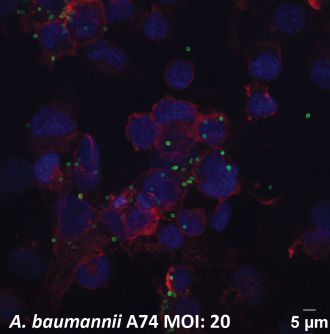
In mouse infection models, the antibody successfully eliminated multidrug-resistant Acinetobacter baumannii, a notorious cause of hospital-acquired pneumonia and bloodstream infections.
“Multidrug resistant Acinetobacter baumannii is a critical threat faced in modern healthcare facilities across the globe,” Professor Goddard-Borger said. “It is not uncommon for infections to resist even last-line antibiotics. Our work serves as a powerful proof-of-concept experiment that opens the door to the development of new life-saving passive immunotherapies.”
Passive immunotherapy involves administering ready-made antibodies to rapidly control an infection, rather than waiting for the individual’s adaptive immune system to respond to the infection. This strategy can be used both therapeutically and prophylactically, which could be deployed to protect vulnerable patients in intensive care units.
Associate Professor Scott said the antibodies also provide a powerful new tool for understanding how bacteria cause disease.
“These sugars are central to bacterial virulence, but they’ve been very hard to study,” he said. “Having antibodies that can selectively recognise them lets us map where they appear and how they change across different pathogens. That knowledge feeds directly into better diagnostics and therapies.”
Over the next five years, the team aims to translate these findings into clinic-ready antibody therapies targeting multidrug resistant A. baumannii. Success would effectively remove the “A” from the ESKAPE pathogens – a milestone in the global fight against antimicrobial resistance.
“This is exactly the kind of breakthrough the new ARC Centre of Excellence is designed to enable,” Professor Payne said. “Our goal is to turn fundamental molecular insight into real-world solutions that protect the most vulnerable people in our healthcare system.”
DOWNLOAD photos of researchers and a copy of the research at this link.
RESEARCH
Tang, A. et al ‘Uncovering bacterial pseudaminylation with pan-specific antibody tools’ (Nature Chemical Biology 2026). DOI: 10.1038/s41589-025-02114-9
DECLARATION
The authors declare no competing interests. Funding was received from the National Health and Medical Research Council; Australian Research Council; National Institutes of Health; the Walter and Eliza Hall Institute of Medical Research; Victorian State Government. Researchers acknowledge support of the Melbourne Mass Spectrometry and Proteomics Facility at the Bio21 Molecular Science and Biotechnology Institute.
All animal handling and procedures were conducted in compliance with the University of Melbourne guidelines and approved by the University of Melbourne Animal Ethics Committee (application ID 29017).
Multimedia






 Australia; NSW; VIC
Australia; NSW; VIC