Media release
From:
New test could predict disease severity in children with rare FOXG1 syndrome
Researchers in Taiwan and Belgium developed a lab-based diagnostic workflow that could help doctors predict the severity of a rare childhood brain disorder called FOXG1 syndrome (a rare neurodevelopmental disorder, like Rett syndrome). The findings could support earlier and more personalized interventions for children with FOXG1 gene mutations.
Published today in Molecular Psychiatry, the study offers the first practical tool to assess whether different FOXG1 gene mutations are likely to cause mild or severe brain abnormalities. FOXG1 syndrome affects children globally with an estimated incidence of 1 in 30,000 live births based on 2020 research by López-Rivera et al. in the journal Brain. The syndrome causes developmental delays, epilepsy, and severe intellectual disability. About 1,200 confirmed cases are documented in global registries, with around 20 families affected across Australia.
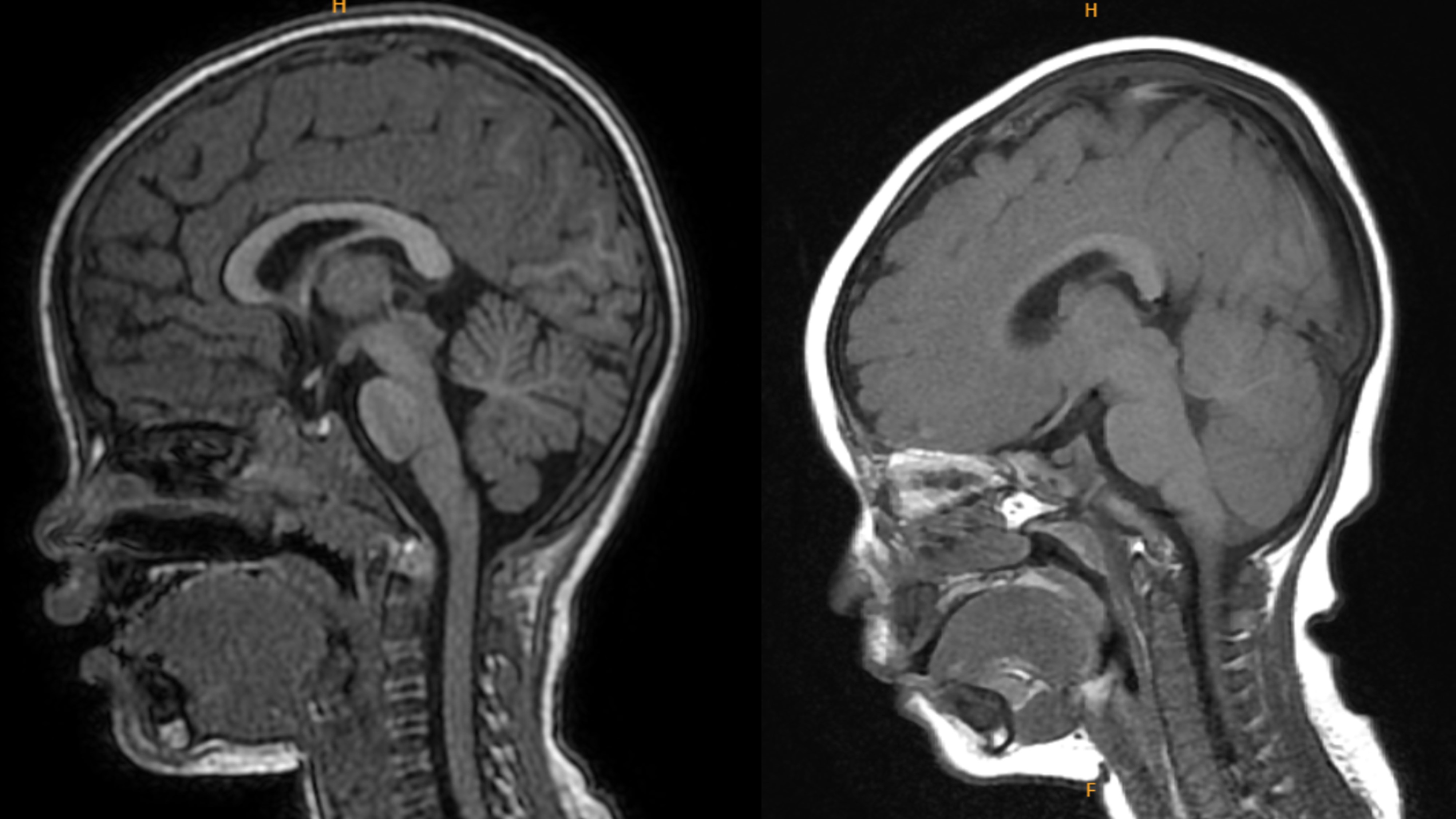
The team analyzed brain imaging and clinical data from 14 individuals diagnosed with FOXG1 syndrome across Europe, North America, Japan, and Taiwan. They then tested the disease-causing variants using three lab tests and successfully predicted brain severity in over 90% of cases. The lab tests include measuring protein expression, checking how FOXG1 regulates key brain development genes, and assessing how human FOXG1 variants affect neuronal migration in mouse embryos.
"This approach helps doctors go beyond simply identifying a mutation, to understanding how dangerous it actually is," said Professor Jin-Wu Tsai from National Yang Ming Chiao Tung University (NYCU), corresponding author of the study. "It's a major step toward precision care for families affected by this genetic disease. For the first time, we can assess the pathogenic impact of specific FOXG1 mutations and predict different levels of severity in patients—critical steps in personalizing care."
The authors say the test may have potential for future prenatal or neonatal diagnostic development and could potentially help parents and clinicians make more informed decisions.
FOXG1 syndrome is caused by spontaneous mutations in the FOXG1 gene, which plays a critical role in early brain development. Symptoms vary widely among patients, and until now, it has been difficult to explain or predict those differences. This can leave families unsure whether to seek further medical advice, how concerned they should be, or what steps to take after a diagnosis. While the study does not address treatment, the authors believe their findings could help address some of these challenges and support future research into targeted therapies for specific FOXG1 variants.
"Rare diseases often leave families with more questions than answers," said Dr. Wang-Tso Lee, Professor and Superintendent at National Taiwan University (NTU) Children's Hospital, co-corresponding author of the study. "We hope this diagnostic tool can help close that gap. By enabling early and accurate prediction of disease severity—even in the prenatal or neonatal period through next-generation sequencing—we can help inform future intervention strategies and better support children and families affected by this devastating disorder."
While the study is limited by a small number of cases and uses specialized lab techniques not yet common in routine testing, the findings provide early evidence that could lead to more personalized diagnostic approaches. The researchers note that broader validation and more accessible testing tools will be needed before this method can be widely adopted in clinical care.



 International
International