News release
From:
1. Genomics: AlphaGenome predicts the impact of DNA variations *PRESS BRIEFING* *IMAGES & VIDEO*
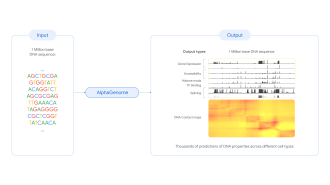
AlphaGenome, a deep learning model that can predict the function of long DNA sequences — up to one million base pairs — is presented in Nature this week. The tool can predict how DNA sequence variations affect different biological processes. AlphaGenome could be used to learn more about genetic diseases, improve genetic testing and inform the development of new treatments.
Genetic variation affects biological processes and can contribute to diseases but understanding how changes to DNA sequences affect their function is challenging. Most changes (around 98%) happen in non-coding regions (parts of DNA that do not code for proteins but can influence how genes are expressed), making it hard to predict the effects. Computational models are needed to solve this problem. Existing methods must make trade-offs between sequence length and the strength of the predictions, but AlphaGenome can make high-resolution predictions across long DNA sequences.
Žiga Avsec, Natasha Latysheva, Pushmeet Kohli and colleagues at Google DeepMind demonstrate the capabilities of AlphaGenome. The model is trained using human and mouse genomes to learn how DNA sequences affect various biological processes. AlphaGenome can simultaneously predict 5,930 human or 1,128 mouse genetic signals that relate to specific functions, such as gene expression, splicing (the cutting and rearranging of genomes) and modification of proteins. The results match or improve on the performance of existing state-of-the-art models in 25 out of 26 evaluations of variant effect predictions. Its strength lies in the ability to make multiple predictions over a range of genetic signals and biological outcomes simultaneously, the authors note.
Further improvements could be made to expand the applications of this tool, such as increasing the species coverage or extending the range of non-coding sequences that the model knows, the authors write. They conclude that AlphaGenome has the potential to improve the understanding of the complex biological outcomes of DNA sequence variation.
***
Please note that an online press briefing for the paper below will take place UNDER STRICT EMBARGO on Tuesday 27th January at 3pm London time (GMT) / 10 am US Eastern Time.
Authors Pushmeet Kohli (Google DeepMind), Žiga Avsec (Google DeepMind), and Natasha Latysheva (Google DeepMind) will discuss the research. This will be followed by a Q&A session.
To attend this briefing you will need to pre-register by following the link in the attachments. Once you are registered, you will receive an email containing the details for the briefing. You will also be provided with the option to save the details of the briefing to your calendar.
Multimedia



 International
International



