News release
From:
Genomes of 24,000 previously unknown microbes revealed by new tools
- Almost all microbial life on Earth remains unknown.
- A new method ‘SingleM’ was developed which detects unknown microbial species.
- ‘Bin Chicken’ rummaged through 700,000 publicly available metagenomics datasets to extract new genomes.
Associate Professor Ben Woodcroft, from QUT’s Centre for Microbiome Research and School of Biomedical Sciences, based at Brisbane’s Translational Research Institute said microbial communities of bacteria and archaea played vital roles in supporting all life on Earth.
“We know gut microbes are essential for digestion and health, but microbes are just as important in the broader environment—from supporting plant growth in soils to producing the oxygen we breathe in oceans,” Professor Woodcroft said.
“Despite decades of research, more than 99 per cent of microbial species remain unknown.
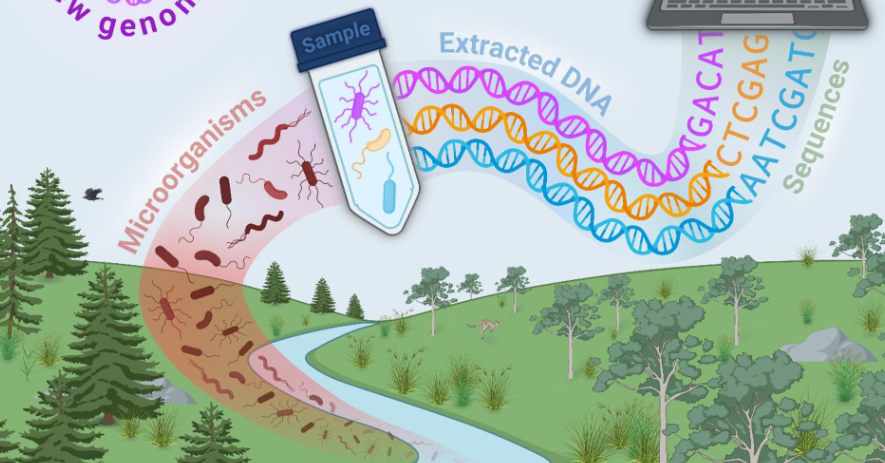
“A key tool in our toolbox for finding new microbial life is ‘metagenomics’, where DNA sequence data is extracted directly from environmental samples.
“The process is very useful because it can be applied almost anywhere in any environment. The challenge lies in processing the data, piecing together the short pieces of DNA we get from metagenomics into full genomes.
“To help close this gap, we developed two software tools to identify and analyse unknown microbes in metagenomics data.”
The first tool, SingleM, quickly scans a microbial sample and identifies what organisms are present—including species that are extremely different from anything seen before.
“The team applied SingleM to more than 700,000 publicly available metagenomics datasets and found that around 75 per cent of the cells in environmental samples belonged to unknown species.”
The second tool, Bin Chicken, dives more deeply into promising samples to reconstruct full genomes from previously uncharacterised microbes.
Lead author and postdoctoral researcher Dr Sam Aroney said Bin Chicken was affectionately named after the Australian white ibis that rummages through garbage bins for morsels of food, “similar to the way our tool rummages through publicly available metagenomic data”.
“Using Bin Chicken, we reconstructed 24,000 new microbial genomes, including several from entirely new branches of the tree of life,” Dr Aroney said.
“These lineages likely evolved before plants and animals and help us better understand life’s early evolution.”
Professor Woodcroft said this expanded genomic catalogue was already revealing new insights into global ecosystems.
“Microorganisms are central to climate change - they’re the largest producers of methane on Earth,” he said.
“We’re now integrating these new genomes into climate modelling and evolutionary studies. They also have broad implications for biotech and health research.”
The research also provided opportunities for the next generation of scientists. QUT undergraduate Joshua Mitchell developed AI methods to predict sample characteristics, such as whether the microbes came from a human host, and has since started a PhD to extend this work.
The full research team includes Associate Professor Woodcroft, Dr Aroney, Rossen Zhao, Joshua Mitchell, Rizky Nurdiansyah, Dr Rhys Newell, and Professor Gene Tyson from QUT; and Mitchell Cunningham and Professor Linda Blackall from the University of Melbourne.
The study, Comprehensive taxonomic identification of microbial species in metagenomic data using SingleM and Sandpiper, is published in Nature Biotechnology alongside a dedicated website https://sandpiper.qut.edu.au.
For more on Bin Chicken see the study Bin Chicken: targeted metagenomic co assembly for the efficient recovery of novel genomes published in Nature Methods.



 Australia; VIC; QLD
Australia; VIC; QLD