News release
From:
Cell biology: Cell maps for the human body (N&V) *PRESS BRIEFING*
Reference cell maps for the human intestine, kidney and maternal–fetal interface (where placenta and maternal cells coexist) are presented in three papers published in Nature this week. These studies are part of a wider package of papers from the Human BioMolecular Atlas Program (HuBMAP) published across the Nature Portfolio journals. The work uncovers new information about how cell types are arranged and how they interact in different human tissues and organs, providing a resource for studying human biology and disease.
In humans, the organization of and interactions between cells determine how organs and tissue function. The HuBMAP initiative aims to map how cells are arranged throughout the human body to help scientists study how cells work and how relationships between cells can affect the health of an individual. To do this, the HuBMAP consortium has been developing tools that aid in assembling spatial maps of cell molecular components, including RNAs, proteins and metabolites, within tissues and organs at a single-cell level. Such tools have been used to generate reference cell atlases for the human intestine, kidney and tissues connected to the placenta.
Michael Snyder, Garry Nolan, William Greenleaf and colleagues studied the human intestine, a complex organ with many different structures and functions (from digestion to supporting the immune system). Eight sections of intestine from nine individuals were analysed, revealing dramatic variations in cell composition across the different regions. The authors identified new subtypes of epithelial cell and found that distinct cell types formed ‘neighbourhoods’, some of which are specifically poised to mediate immune responses. The findings reveal the complex, varied cell composition that contributes to the functioning of this organ.
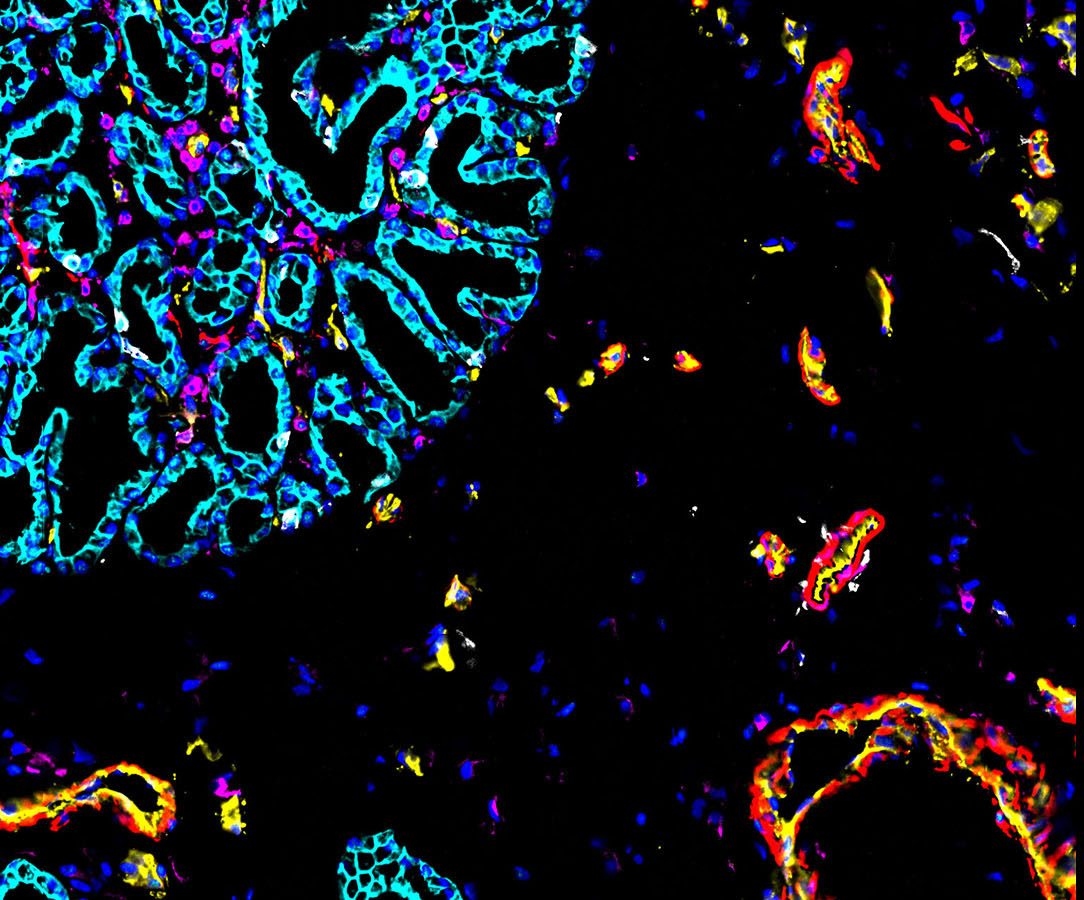
Sanjay Jain, Matthias Kretzler, Kun Zhang, Tarek M. El-Achkar, Pierre C. Dagher, Michal T. Eadon and colleagues examined cells in 45 healthy and 48 diseased human kidneys. Damage to these organs can trigger changes in kidney cells that ultimately affect kidney function. The authors produce a single cell and spatial atlas of 51 main cell types across different regions of the kidney. They also identify cellular states and neighborhoods of kidney immune, stromal and epithelial cells that are altered by acute or chronic injury, including states related to whether repair pathways are successful or defective.
Michael Angelo, Shirley Greenbaum and colleagues constructed a map of the human placenta during the first half of pregnancy. They analysed around 500,000 cells and 588 arteries from 66 samples of the human maternal–fetal interface (where maternal and placental cells cooperate to support the fetus). Specifically, they looked at the interface between the placenta and uterus, where maternal arteries are remodelled to deliver blood to the fetus. The maps span different stages of development (6–20 weeks gestation) and identify interactions between placental and immune cells; the latter discovery sheds light on how the maternal immune cells support the coexistence of the distinct maternal and fetal cells.
“The three HuBMAP atlases […] have the potential to advance our understanding of disease, by defining the spatial location of cell states linked to disease,” write Roser Vento-Tormo and Roser Vilarrasa-Blasi in an accompanying News & Views article. They anticipate the generation of more atlases in other tissues but note that even more tests must be carried out on more samples to “establish robust associations between cellular organization and function in health and disease”.
The full collection will be available upon publication here.
**Please note that an online press briefing for the paper below will take place UNDER STRICT EMBARGO on Tuesday 18 July at 4 pm London time (BST) / 11 am US Eastern Time**
Authors Michael Snyder, Sanjay Jain, Michael Angelo, Katy Borner and Richard Conroy will discuss the research. This will be followed by a Q&A session.
To attend this briefing you will need to pre-register by following the link here. Once you are registered, you will receive an email containing the details for the briefing. You will also be provided with the option to save the details of the briefing to your calendar.



 International
International