News release
From:
Developmental biology: Synthetic mouse embryos generated *UK SMC BRIEFING*
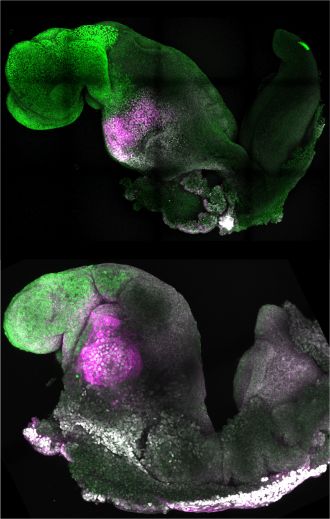
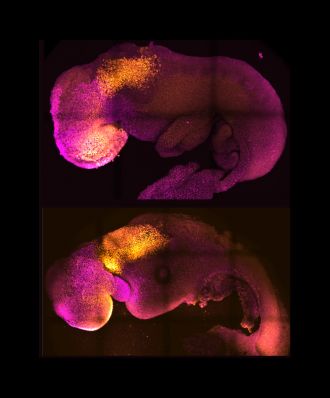
The generation of synthetic, stem cell-derived mouse embryos is described in a paper published in Nature this week. The embryo model copies the stages of natural mouse embryo development that take place up to day 8.5 post fertilization, including the establishment of defined regions of the brain, a neural tube and a beating heart-like structure. In addition, the model can replicate the consequences observed in natural mouse embryos from the knockout of a gene. The finding presents the possibility of using this model to understand factors that regulate the early stages of development, without the need for experimental animals.
Embryonic stem cells have the ability to form embryo-like structures in the laboratory. However, these models do not completely mimic all phases of development; for example, they do not fully recapitulate a process called neurulation (the formation of the neural tube, which will eventually differentiate into the brain and spinal cord).
Magdalena Zernicka-Goetz and colleagues assembled stem-cell derived mouse embryos in the laboratory using a combination of embryonic stem cells, trophoblast stem cells and inducible extraembryonic endoderm stem cells, all from mice. The resulting ETiX-embryoid model was able to develop beyond neurulation to the equivalent of 8.5 days post fertilization in a natural mouse embryo and established all the brain regions, a neural tube, a beating heart and a gut tube. The authors note that the model was able to achieve this through self-organization of the stem cell types, without the need for external signalling cues. In further experiments, the authors were able to demonstrate that knockout of the gene Pax6 — involved in the development of the eyes and other sensory organs — in their embryoid model resulted in similar effects to those seen in natural Pax6 knockout mouse embryos.
Zernicka-Goetz and co-authors conclude that the ETiX-embryoids provide a physiological relevant model of embryo development and present a new opportunity to study mechanisms of development and disease.
An online press briefing, hosted by the UK Science Media Centre, will take place on Wednesday 24 August at 4pm London time (BST).
In order to attend the press briefing you will need to pre-register by following the link.
Multimedia




 International
International



