News release
From:
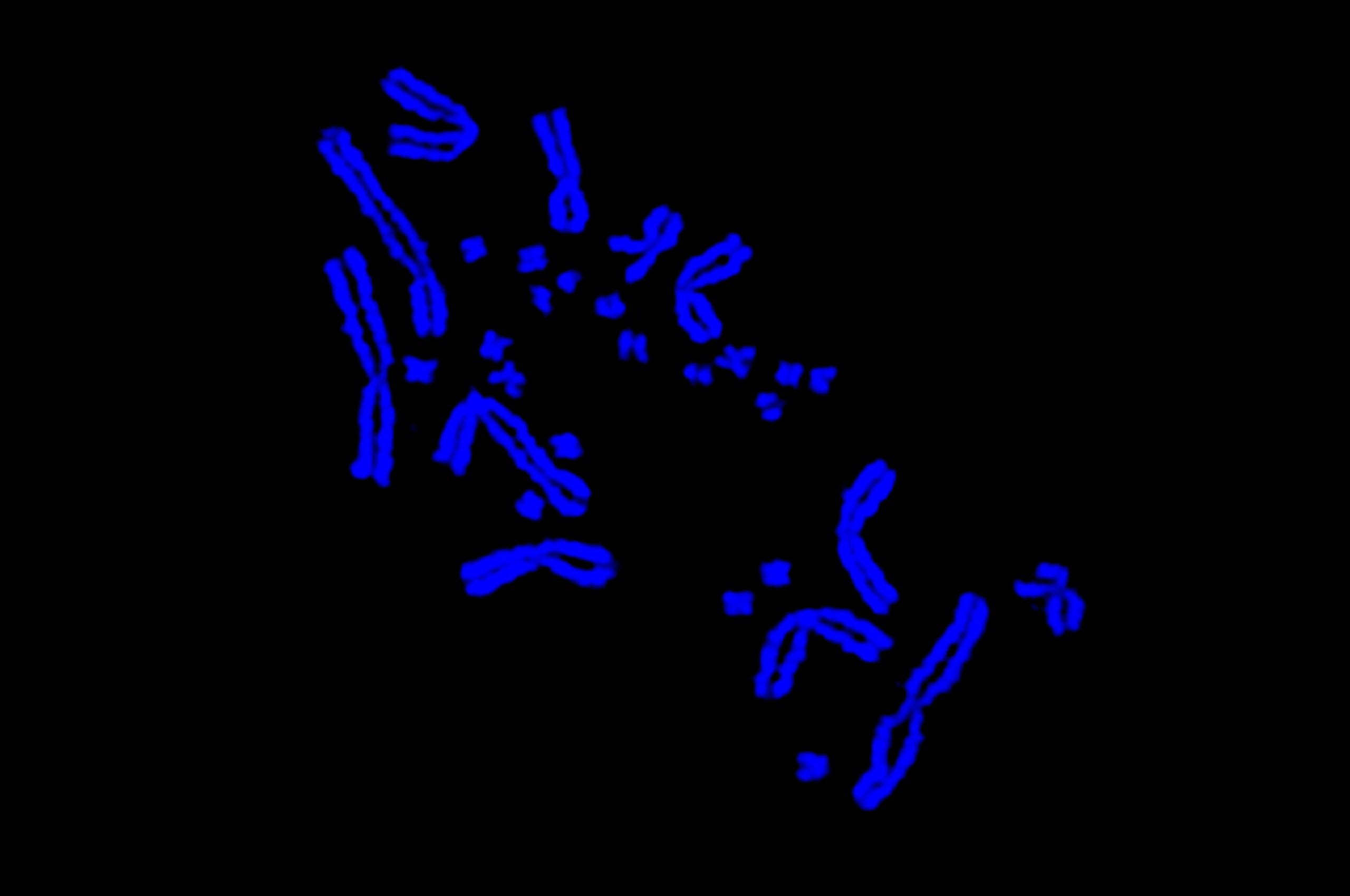
Scientists have discovered that tiny ‘microchromosomes’ in birds and reptiles, initially thought to be specks of dust on the microscope slide, are linked to a spineless animal ancestor that lived 684 million years ago.
They prove to be the building blocks of all animal genomes, but underwent “dizzying rearrangement” in mammals, including humans.
Publishing findings today in PNAS, a team led by Professor Jenny Graves at La Trobe University, made the discovery by lining up the DNA sequence of microchromosomes that huddle together in the cells of birds and reptiles.
When these little microchromosomes were first seen under the microscope, scientists thought they were just specks of dust among the larger bird chromosomes, but are actually proper chromosomes.
Professor Graves said that, using advanced DNA sequencing technology, scientists can at last sequence microchomosomes end-to-end.
“We lined up these sequences from birds, turtles, snakes and lizards, platypus and humans and compared them,” Professor Graves said.
“Astonishingly, the microchromosomes were the same across all bird and reptile species. Even more astonishingly, they were the same as the tiny chromosomes of Amphioxus – a little fish-like animal with no backbone that last shared a common ancestor with vertebrates 684 million years ago,” Professor Graves said.
Professor Graves said in marsupial and placental mammals these ancient genetic remnants are split up into little patches on our big, supposedly normal, chromosomes.
“The exception is the platypus genome, in which the microchomosomes have all fused together into a few large blocks that reflect our oldest mammal ancestor,” Professor Graves said.
Professor Graves said the findings highlight the need to rethink how we view the human genome.
“Rather than being ‘normal’, chromosomes of humans and other mammals were puffed up with lots of ‘junk DNA’ and scrambled in many different ways,” Professor Graves said.
“The new knowledge helps explain why there is such a large range of mammals with vastly different genomes inhabiting every corner of our planet.”
Professor Graves led a team of researchers from the Australian National University, the University of Canberra and University of New South Wales, as well as international collaborators.
“It was exciting to work with young people with the skills to analyse complex genome data,” Professor Graves said.
The study is Professor Graves’ inaugural article in PNAS after her election as an International Member of the prestigious United States National Academy of Science in 2019.
One of the highest honours a scientist can receive, her election recognised her distinguished research in genetics and genomics. There are only 24 other Australian scientists who currently hold this honour.
Professor Jenny Graves AO
Professor Graves is renowned for her pioneering investigation into the genetics of sex, and her work mapping the genomes of several Australian animals.
In 2017 she became the first individual woman to be awarded Australia’s highest honour for the sciences, the Prime Minister’s Prize for Science.
Professor Graves is now participating in the Earth Biogenome Project, a 10-year global effort she helped launch in 2018 to sequence the genetic code of all 1.5 million known animal, plant, protozoan, and fungal species on Earth.
Multimedia




 Australia; VIC
Australia; VIC