Media Release
From: Cell PressPeer-reviewed Experimental Study Animals/Mice
Researchers identify novel antibiotics using machine learning
Despite growing concerns about antibiotic resistance, the discovery of new antibiotic drugs has slowed as new molecules become increasingly difficult to identify. But in a paper publishing February 20 in the journal Cell, researchers demonstrate a method to uncover new antibiotics quickly and efficiently through machine learning.
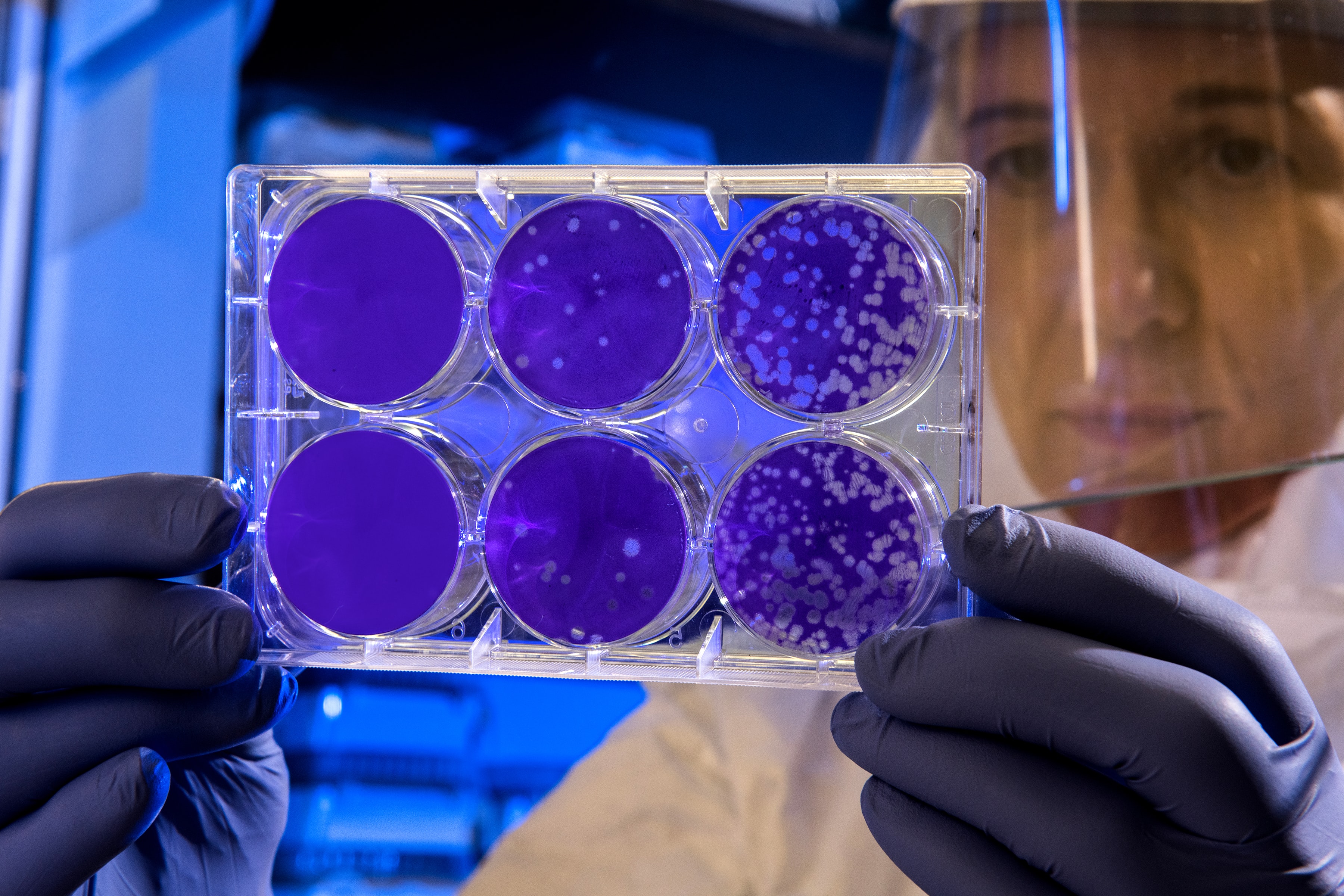
James Collins, a synthetic biologist at MIT, and his team trained a deep neural network to identify possible antibiotic molecules using compounds known to suppress E. coli growth; the network then used these data to examine other molecules in existing chemical libraries and predict their likelihood to suppress growth of bacteria. The researchers found that nearly 50% of those compounds they prioritized to test showed some effectiveness in vitro in limiting E. coli proliferation. One compound in particular, halicin, is structurally divergent from conventional antibiotic molecules and uses a mechanism that’s uncommon in clinical antibiotics to fight a wide range of human pathogens in mice, including Clostridioides difficile and Acinetobacter baumannii.
While these vast libraries of molecules have been available as potential sources of new antibiotics, researchers haven’t had an efficient way before now to curate them to identify promising candidates for experimental testing. But if widely adopted, deep neural networks could save both time and resources in the search for new, unique antibiotics—something, the authors say, that is necessary in part because of a lack of economic incentives for their discovery. They write, “deep learning approaches could enable us to outpace the emergence of multidrug-resistant pathogens and usher in the Second Age of Antibiotics.”